June 11, 2016 Harvard and the Broad Institute Deliver Precision Medicine Advancement through Crowdsourcing
In April of this year, Harvard and Topcoder announced an ambitious series of data science challenges focused on driving advancements in precision medicine. The hope, that with the use of crowdsourcing competitions, Harvard, with their partners at the Broad Institute, and funding from the Kraft Foundation, could optimize certain key algorithms crucial to research driving the burgeoning field of study.
Together, Harvard and the Broad Institute created a crowdsourcing program to break data bottlenecks through prize-based challenges in order to pursue the research goals of the HBS and the Crowd Innovation Laboratory (CIL) for the advancement of precision medicine.
What is Precision Medicine?
In broad strokes, it’s the practice of tailoring medical treatment – decisions, drugs, therapies and all – to an individual or sub-group. The field shows incredible promise.
Crowdsourcing Competitions Kick-Off on Topcoder
The purpose of DNAS1 (DNA Sequencing 1) was to look for an algorithm that aligns multiple DNA sequences to a reference DNA for a simpler case, when there are only minor differences between the reference DNA and the DNA the sequences are originated from. The task was to align the sequences fast, align them right, and test the alignment position for possible redundancies.
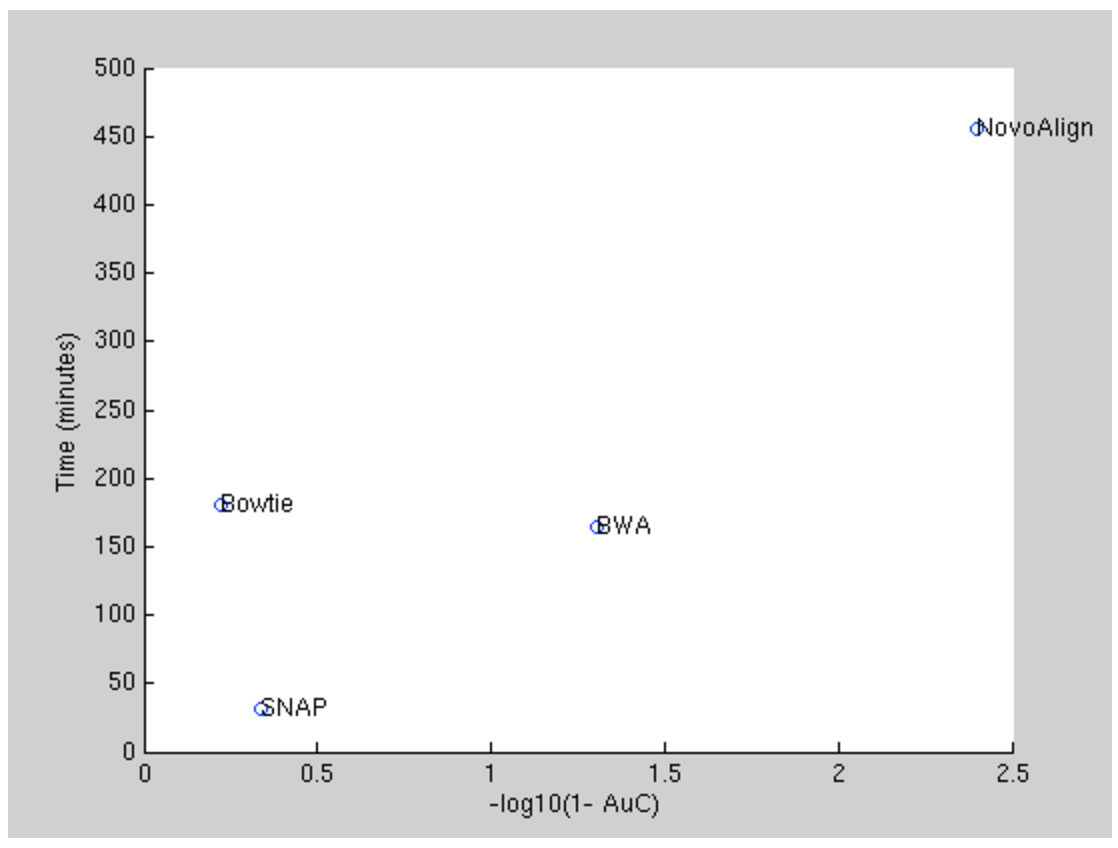
By now there have been multiple attempts on sequence alignment algorithms. The four most popular algorithms currently are: Novoalign, BWA-MEM, Bowtie, and SNAP. Below we compare those four on the large alignment test, similar to the one we used in the DNAS1 contest.
According to the best balance between the accuracy (X-axis) and computational time (Y-axis), the BWA-MEM algorithm was selected to represent the state of the art benchmark for the contestants.
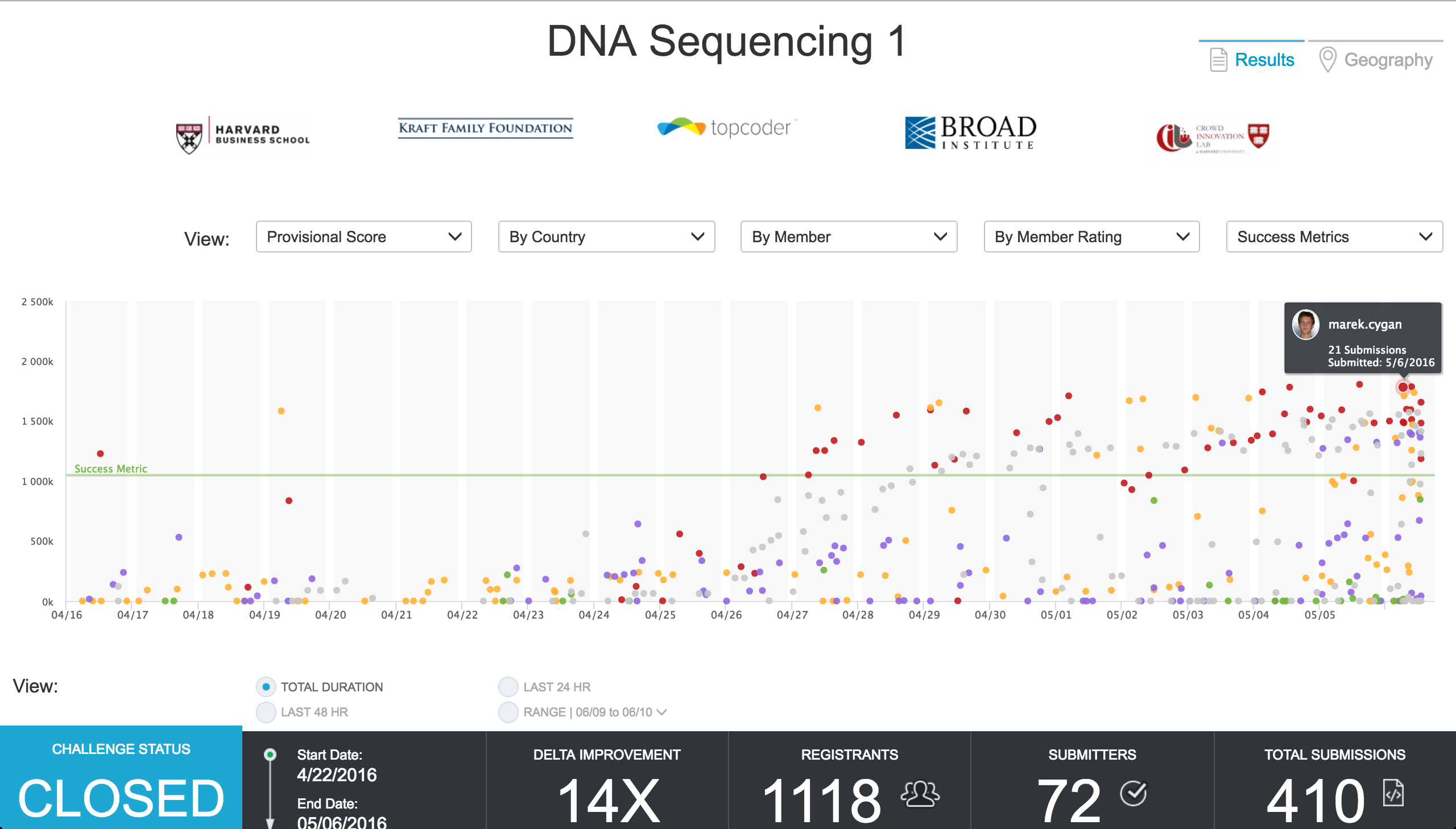
After establishing ground truth data and successfully testing the contest data, the benchmark algorithm (for comparing results), the objective algorithm (which assigns scores to solutions), and all supporting software, the series of crowdsourcing challenges kicked-off in mid-April with Topcoder Marathon Match DNAS1 going live on April 16th.
This crowdsourcing challenge drew robust participation from the 1 million+ member community of Topcoder.
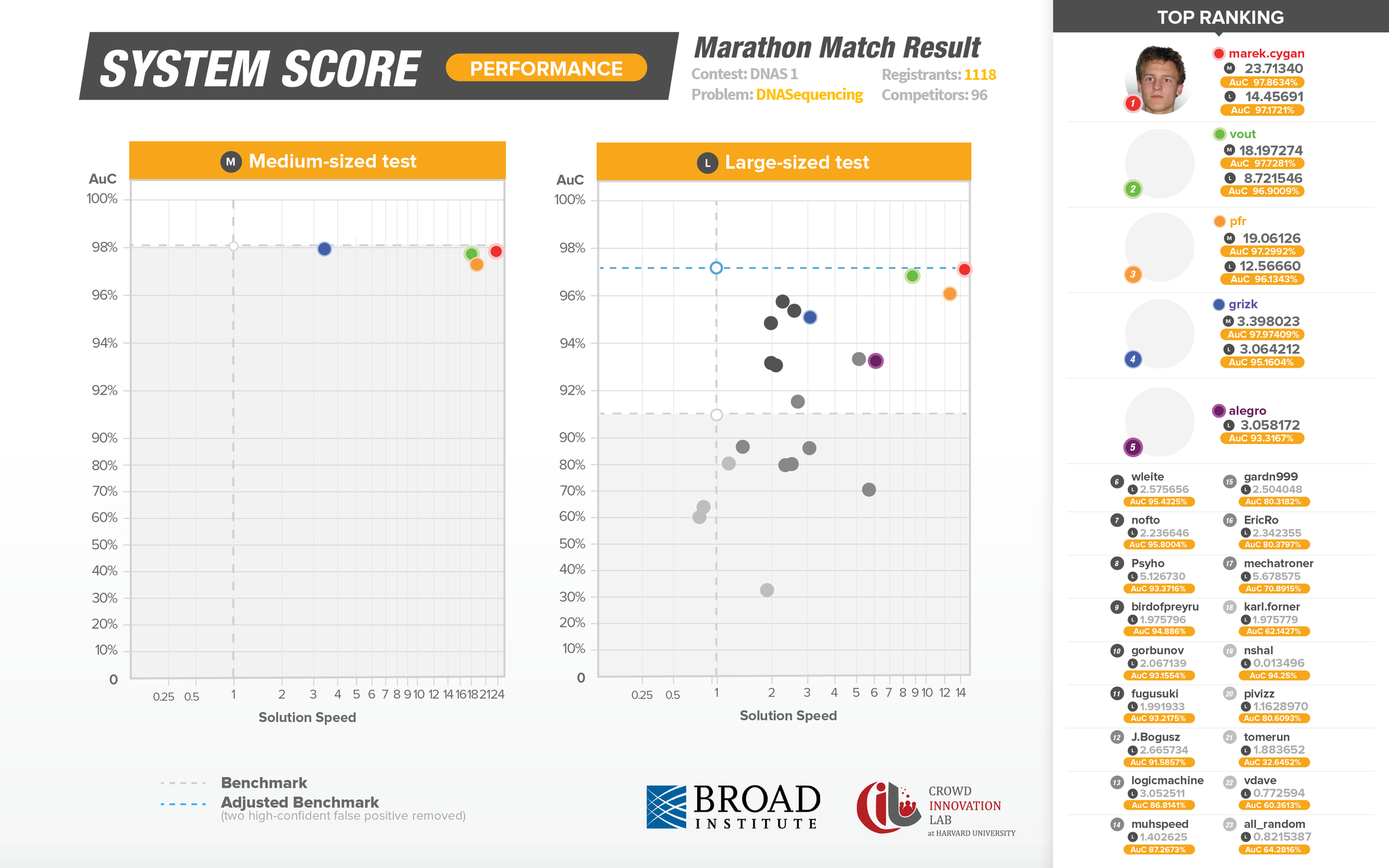
1,118 Topcoder members registered for this precision medicine focused match. Of them, 96 competitors ran test cases and 72 members of the Topcoder community actively competed on this challenge. The 72 members submitted an astounding 410 unique algorithmic solutions over the course of the 3-week long crowdsourcing competition. This means the average competitor submitted ~ 5.7 unique algorithms each. The first-place Topcoder competitor, marek.cygan of Poland, submitted 21 unique algorithmic solutions in his winning effort.
In Data Science, crowdsourcing provides organizations a unique vehicle to help achieve extreme value solutions. Harvard research has continually demonstrated that no other methodology can harness that level of focused solutioning while applying game mechanics to drive individuals to perform repeatedly within the competition. Due to the solution sample size crowdsourcing challenges can garner, extreme value solutions can be achieved.
So, how did the Topcoder community perform?
Crowdsourcing Results from Precision Medicine Data Science Challenge DNAS1
The following data and graphs were created by the Crowd Innovation Lab at Harvard. CIL partners with Topcoder on running ambitious data science crowdsourcing challenges. The CIL at Harvard provides data science experts to determine which challenges to run, the creation of all phases of testing, the launching and executing of the challenge, and the post-challenge analysis like what you see below.
The infographic below represents the provisional scores tallied on the Topcoder platform as the submissions were coming in live.
Some interesting plot points to examine:
- On day 1, Topcoder member pfr scored significantly higher than the agreed upon Acceptable Outcome
- By day 4 of the competition, Topcoder member vout had already scored higher than the established Highest Expectations barometer
- Eventual 1st-place winner, marek.cygan scores low in the opening days of the challenge as he probes the properties of the scoring function.
- As the competition advances, the volume of participants and submissions swells through the finish line
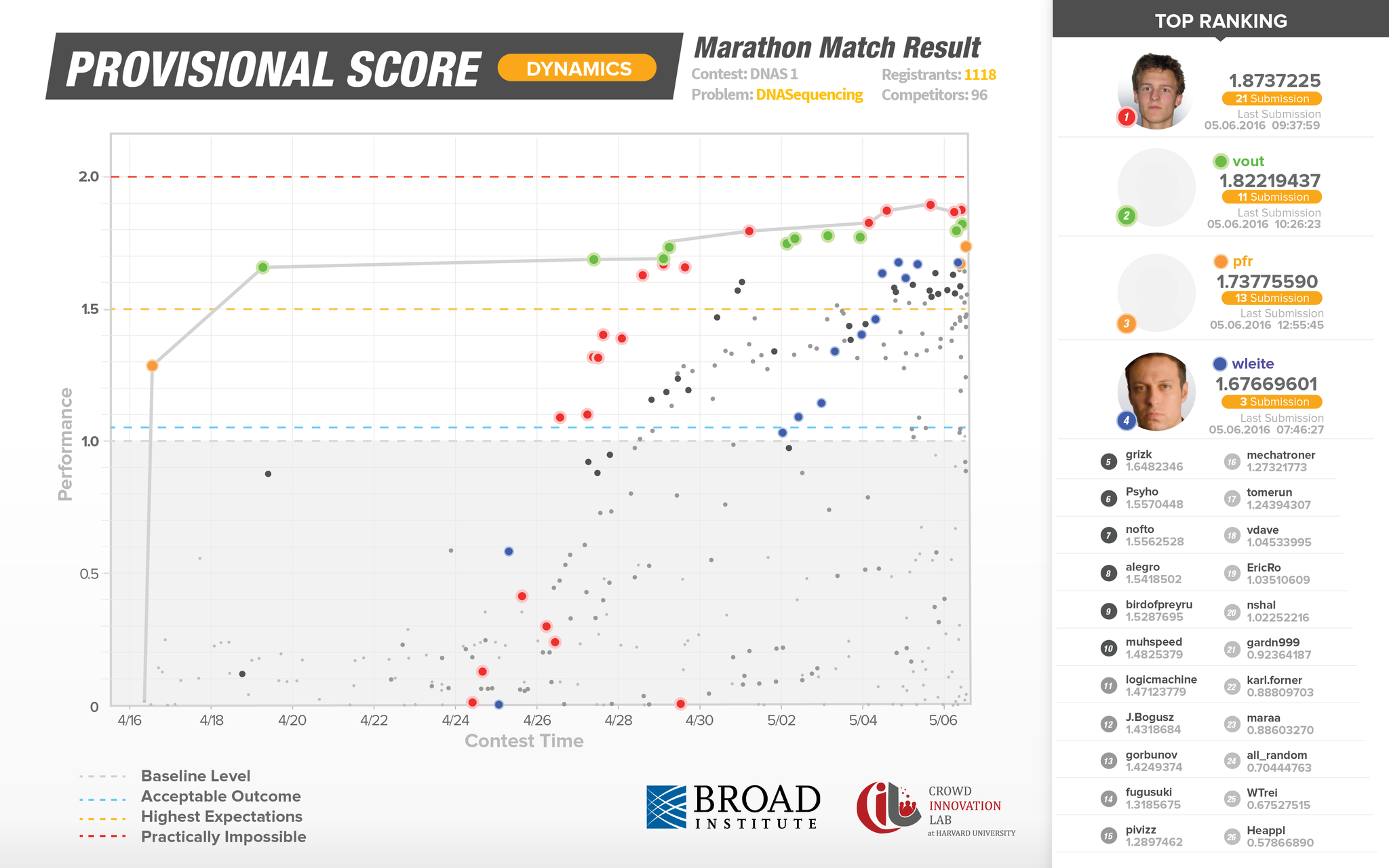
This next graphic represents System Scoring and takes place after the live competition has ended. This second series of testing solutions is crucial in identifying false-positives and as a result is a more accurate reflection of submission performance. The Y axis represents solution accuracy, while the X axis represents performance, in this case speed.
Though several Topcoder members delivered outstanding solutions, the top solution from marek.cygan, performed 14x faster and at the same level of accuracy as the existing in-use algorithmic solution.
Another graph to demonstrate the extreme value solution curated through this Topcoder crowdsourcing competition is the below. While recognizing a 14x improvement to algorithmic speed is important, this graph below explains what the 14x improvement means in terms of time.
For the large-alignment test: The benchmark – existing algorithmic solution – had a solution runtime of 6,865 seconds or slightly over 114 minutes. The winning solution from the crowdsourcing competition, delivered by marek.cygan, has a solution runtime of 475 seconds, or slightly under 8 minutes.
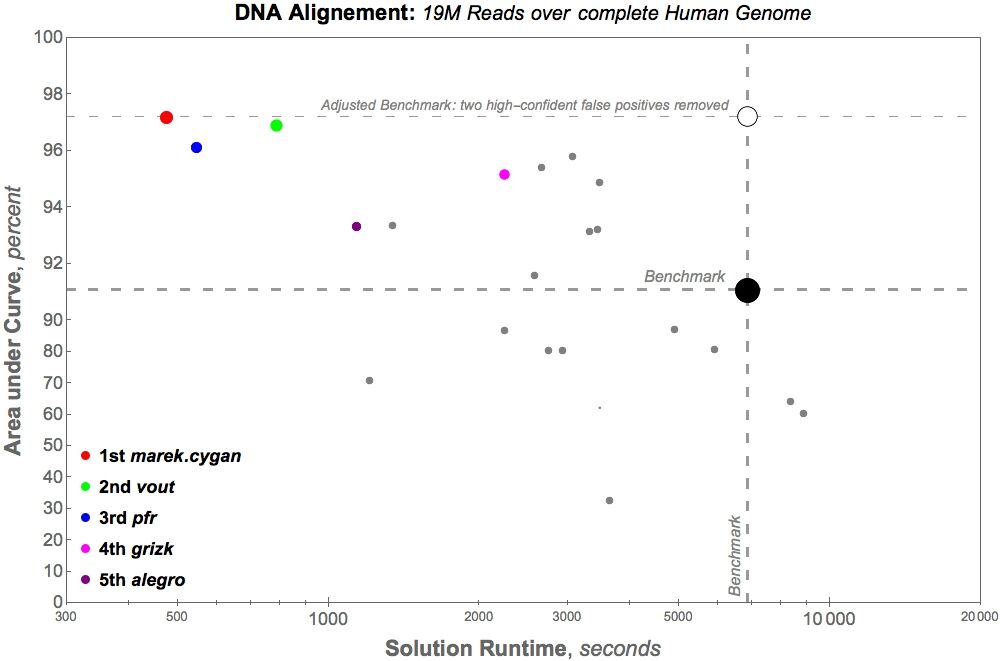
Why does this speed-up matter? Researchers of all kinds desire to run more experiments and to analyze more data. With more optimized algorithms, like the kind delivered through Topcoder Marathon Match DNAS1, researchers can do a larger volume of their work in reduced time frames. These gains can have a huge impact and help lead to critical discoveries faster.
From 1st place winner – marek.cygan, describing his approach to this unique challenge.
“I should say that this was the 1st time I have approached a computational biology problem, in particular I have no prior knowledge whatsoever about DNA sequencing. I decided to enter this challenge as it appeared to me as a nice mix of algorithm design, code optimization and (possibly) machine learning. My solution is self contained i.e., I did not use any special libraries nor
third-party code.”
On the objective function (which assigns scores to competitor submissions):
“I admit I did not have a good intuition on how the objective function works for the first few hours spent on this problem. Later, I noticed that it is crucial not to have bad answers with high confidence. In particular a single misclassified query can cost 10 – 20% drop in the objective function, which is harsh, but reasonable given the nature of the problem… …Moreover, I realized that to get the best tradeoff between the accuracy and speed parts of the score, for my approach at least, it is very important to have a very fast program so I spent significant amount of time on optimizing code.”
Continuing the Series of Precision Medicine Crowdsourcing Challenges
With the first challenge in the precision medicine series now done, focus will turn to CMAP1 or Connectivity Map Challenge #1.
 From the website linked above:
From the website linked above:
CMAP is a collection of genome-wide transcriptional expression data from cultured human cells treated with bioactive, small molecules and simple pattern-matching algorithms. When these elements are brought together, the results enable the discovery of functional connections between drugs, genes and diseases through the transitory feature of common gene-expression changes.
For this contest, the goal is to maximize the accuracy of the inferred gene expression values, while minimizing the number of the measured gene expressions. Results of this contest will further expand research horizons for computational biologists and scientists who seek to find drugs that cure diseases.
If you’re interested in competing in Topcoder Marathon Match CMAP 1 – click here. Also we are hosting an educational webinar Wed. June 15th with an expert from the Broad Institute who will be answering your questions live! Don’t miss this chance to gain an advantage in the upcoming Topcoder match – click here.
From our partners at the Crowd Innovation Lab at Harvard and the Broad Institute, we want to thank all Topcoder members who participated in this remarkable data science crowdsourcing competition. We will continue to provide blog posts on the progress of this series, so please subscribe to the Topcoder Blog to follow the news.