Downloads:
This text attempts to explain in details how the tests are generated. It follows the Python code closely and clarifies what each specific part of the code does.

First, we load the data from 'initialCounts.csv' file. This data will be used to generate the initial counts (before application of selective pressure) for each library member. Out of 2 columns that you can see in the file, only 'ref_input' is actually used.
full_df = pd.read_csv('initialCounts.csv', index_col=None)
full_df.columns = pd.Index(['peptide', 'input'])
Next, we load the test data generation parameters:
params = pd.read_csv('parameters_training.csv')
The code below iterates through all rows of 'parameters_training.csv' and generates a test case for each row. Variable 'output' will be used for storing the test case data.
for (garbage,p) in params.iterrows():
sys.stdout.write("%s\n" % p['id'])
sys.stdout.flush()
output = {}
# actual generation
The next piece of code generates fitness values for each library member:
def generate_lognormal(N, mu, sigma):
return np.random.lognormal(mu, sigma, N)
def generate_uniform(N, domain):
return np.exp(np.random.uniform(-domain, domain, N))
def generate_w(p):
if p['distribution'] == 'lognormal':
return generate_lognormal(p['N'], p['mu'], p['sigma'])
elif p['distribution'] == 'uniform':
return generate_uniform(p['N'], p['domain'])
def centered(x):
return np.exp(np.log(x) - np.mean(np.log(x)))
....
output['w'] = centered(generate_w(p))
Note that p[name] allows to access the value of the test case parameter named name.
In order to generate fitness values, different distributions can be used. All training data files use either log-uniform (generated by generate_uniform) or log-normal (generated by generate_lognormal) distributions. Method generate_w calls one of two specific distribution generation methods with proper parameters depending on the value of parameter distribution. Method centered processes fitnesses so that arithmetic average of their natural logarithms becomes equal to 0.
It's important that submission and system tests can feature different (secret) distributions for fitness values. You can still assume that fitnesses will be post-processed with centered method after being generated, like it happens for training test cases.
Now let's generate the initial counts for each library member:
def normalized(x):
return np.float_(x) / np.sum(x)
....
random_idxs = random.sample(xrange(full_df.shape[0]), p['N'])
df = full_df.ix[random_idxs]
output['X0'] = np.random.multinomial(p['N'] * p['n0'], normalized(np.array(df['input']))) + 1
First, we choose N random rows from 'initialCounts.csv' and consider the values of 'ref_input' at those rows. These values are scaled so that their sum becomes equal to 1 (using normalized method) and passed as probabilities to multinomial distribution generator. We draw N * n0 samples from this multinomial distribution into column of the test case called X0. Finally, each element of X0 is increased by 1, so that none of them are equal to 0.
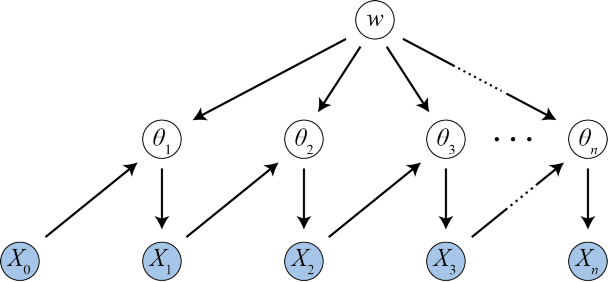
So, now we have fitness values and initial counts. Our last goal is to generate the final counts (after application of selective pressure):
for i in range(1,4):
if pd.isnull(p['n%i' % i]):
break
output['theta%i' % (i)] = np.random.dirichlet(normalized(output['X%i' % (i-1)] * output['w']) * np.sum(output['X%i' % (i-1)]))
output['X%i' % (i)] = np.random.multinomial(p['N'] * p['n%i' % i], output['theta%i' % i]) + 1
The generation works in one or several phases (at most 3). The i-th phase is applied if and only if the value of parameter ni is not 'nan'. Here's what exactly happens:
Once the test case is generated, we can save it into a file:
output_df = pd.DataFrame(output)
output_df.to_csv("%s.csv" % p['id'], index=False)
For all training data files all the columns Xi, thetai and fitness values are known to you. However, for submission and system test cases you will be given only the columns Xi and your task will be to estimate the fitnesses as precisely as possible.